A colleague of mine asked me if I was aware of an analogous format for the global FAO fisheries statistical areas. After some search I found indeed that FAO provides these statistical areas in a Google Earth readable format. But a little cumbersome for my taste. Luckily, the shapefiles are easily available at the FAO site. So here is my take on things:
################################################################################ # FAO statistical areas in Google Earth format require(rgdal) require(plotKML) setwd("~/Dropbox/blogspotScripts/20120701FAOareas/") # your 'native' directory url <- 'http://www.fao.org/geonetwork/srv/en/resources.get?id=31627&fname=fa_.zip&access=private' download.file(url,destfile="~/Downloads/tmp.zip") # your 'native' directory unzip("~/Downloads/tmp.zip") # your 'native' directory fao <- readOGR(".","fa_") setwd("~/Dropbox/Public/") # your 'native' directory plot(fao) # a peek preview in R # here one could simply do, via function from the plotKML package kml(fao,file="tmpFAO.kml") # open your tmp.kml in GE to have a view # but that stuff is no good # to put some order to things, etc. names(fao) <- tolower(names(fao)) dat <- as.data.frame(fao) i <- data.frame(str_locate(fao$f_subarea,"_")) i <- !is.na(i[,1]) fao$f_subarea[i] <- str_sub(fao$f_subarea[i],2) fao$f_division[i] <- str_sub(fao$f_division[i],2) fao$f_subdivis[i] <- str_sub(fao$f_subdivis[i],2) fao$name <- ifelse(!is.na(fao$f_subunit),fao$f_subunit, ifelse(!is.na(fao$f_subdivis),fao$f_subdivis, ifelse(!is.na(fao$f_division),fao$f_division, ifelse(!is.na(fao$f_subarea),fao$f_subarea, fao$f_area)))) fao <- as(fao,"SpatialLinesDataFrame") fao$id <- as.numeric(fao$f_area) area <- sort(unique(fao$f_area)) # put the stuff in order of the major areas # again some function of the plotKML packages put in use kml_open("fao.kml") # filling up the russian doll for (i in 1:length(area)) { tmp <- fao[fao$f_area %in% area[i],] kml_layer.SpatialLines(tmp,labels=tmp$name,folder.name = area[i]) } kml_close("fao.kml") kml_compress("fao.kml") # create a compressed (KMZ) file
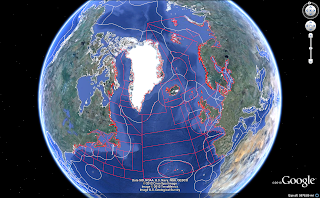
Did not quite get want I really wanted (plotKML is really in its infancy, just released to cran last week. But a very promising package). So played a little around (non reproducible code) to end up, preliminary with this, the following being a snapshot: